Extended: Plotting Phenotypic Correlation by Degree of Relatedness
S. Mason Garrison
2026-02-07
Source:vignettes/articles/v31_phenotypicbydegree.Rmd
v31_phenotypicbydegree.RmdThis vignette demonstrates how to visualize phenotypic correlation by
degree of relatedness using the ggPhenotypeByDegree
function from the ggpedigree package. This function is
particularly useful for analyzing and visualizing the relationship
between phenotypic traits and genetic relatedness in pedigree data. It
extends the basic examples found in the main package documentation.
Click to expand pedigree setup
library(ggpedigree)
library(BGmisc)
library(data.table)
# Load the example data
data("redsquirrels")
library(dplyr)
# library(broom) # for tidy()
library(purrr) # for map_* helpers
# Filter for the largest family, recode sex if needed
ped_filtered <- redsquirrels_full %>%
recodeSex(code_female = "F") %>%
filter(famID == 160)
kin_degree_max <- 12 # maximum degree of relatedness to consider
# Calculate relatedness matrices
add_mat <- ped2add(ped_filtered, isChild_method = "partialparent", sparse = TRUE)
mit_mat <- ped2mit(ped_filtered, isChild_method = "partialparent", sparse = TRUE)
cn_mat <- ped2cn(ped_filtered, isChild_method = "partialparent", sparse = TRUE)
df_links <- com2links(
writetodisk = FALSE,
ad_ped_matrix = add_mat,
mit_ped_matrix = mit_mat,
cn_ped_matrix = cn_mat,
drop_upper_triangular = TRUE,
gc = FALSE
)
dataRelatedPair_merge <- df_links %>%
left_join(ped_filtered %>% select(personID, lrs, ars_n),
by = c("ID1" = "personID")
) %>%
rename(
lrs_k1 = lrs,
ars_n_k1 = ars_n
) %>%
left_join(ped_filtered %>% select(personID, lrs, ars_n),
by = c("ID2" = "personID")
) %>%
rename(
lrs_k2 = lrs,
ars_n_k2 = ars_n
)
# double enter
dxlist <- c(
"ID1", "ID2", # intentional ordering
"addRel", "mitRel",
"cnuRel",
names(dataRelatedPair_merge)[endsWith(names(dataRelatedPair_merge), "_k2")],
names(dataRelatedPair_merge)[endsWith(names(dataRelatedPair_merge), "_k1")]
)
kin_degrees <- 0:12
rel_vals <- 2^(-kin_degrees)
rel_bins <- purrr::map_chr(rel_vals, ~ paste0("addRel_", .x))
rel_labels <- c("addRel_1+" = "addRel_1+")
# bin assignment function
assign_addRel_bin <- function(rel) {
for (val in rel_vals) {
upper <- val * 1.1
lower <- val * 0.9
if (!is.na(rel) && rel >= lower && rel <= upper) {
return(paste0("addRel_", val))
} else if (!is.na(rel) && rel > upper) {
return(paste0("addRel_+", val))
}
}
if (!is.na(rel) && rel > 2^0 * 1.1) {
return("addRel_1+")
}
if (!is.na(rel) && rel == 0) {
return("addRel_0")
}
return(NA_character_)
}
dataRelatedPair_merge <- data.table::rbindlist(
list(
dataRelatedPair_merge,
dataRelatedPair_merge[, dxlist]
),
use.names = FALSE
) %>%
mutate(addRel_bin = vapply(addRel, assign_addRel_bin, character(1))) %>%
mutate(addRel_factor = factor(addRel_bin, levels = c("addRel_1+", rel_bins, paste0("addRel_+", rel_vals), "addRel_+0", "addRel_0"))) %>%
select(-addRel_bin)
result <- dataRelatedPair_merge %>%
group_by(addRel_factor, mitRel, cnuRel) %>%
summarise(
n_pairs = n() / 2, # divide by 2 to account for double counting
lrs_cor_test = list(tryCatch(cor.test(lrs_k1, lrs_k2, use = "pairwise.complete.obs"),
error = function(e) NULL
)),
ars_n_cor_test = list(tryCatch(cor.test(ars_n_k1, ars_n_k2, use = "pairwise.complete.obs"),
error = function(e) NULL
)),
addRel_mean = mean(addRel, na.rm = TRUE),
addRel_sd = sd(addRel, na.rm = TRUE),
addRel_min = min(addRel, na.rm = TRUE),
addRel_max = max(addRel, na.rm = TRUE),
.groups = "drop" # eliminates the need for ungroup()
) %>%
## unpack the two cor.test() objects -------------------------------
mutate(
# ---- LRS pair ----
cor_lrs = map_dbl(lrs_cor_test, ~ if (is.null(.x)) NA_real_ else .x$estimate),
cor_lrs_stat = map_dbl(lrs_cor_test, ~ if (is.null(.x)) NA_real_ else .x$statistic),
cor_lrs_p = map_dbl(lrs_cor_test, ~ if (is.null(.x)) NA_real_ else .x$p.value),
cor_lrs_df = map_dbl(lrs_cor_test, ~ if (is.null(.x)) NA_real_ else .x$parameter),
cor_lrs_ci_lb = map_dbl(lrs_cor_test, ~ if (is.null(.x)) NA_real_ else .x$conf.int[1] * sqrt(2)),
cor_lrs_ci_ub = map_dbl(lrs_cor_test, ~ if (is.null(.x)) NA_real_ else .x$conf.int[2] * sqrt(2)),
# ---- ARS‑n pair ----
cor_ars_n = map_dbl(ars_n_cor_test, ~ if (is.null(.x)) NA_real_ else .x$estimate),
cor_ars_n_stat = map_dbl(ars_n_cor_test, ~ if (is.null(.x)) NA_real_ else .x$statistic),
cor_ars_n_p = map_dbl(ars_n_cor_test, ~ if (is.null(.x)) NA_real_ else .x$p.value),
cor_ars_n_df = map_dbl(ars_n_cor_test, ~ if (is.null(.x)) NA_real_ else .x$parameter),
cor_ars_n_ci_lb = map_dbl(ars_n_cor_test, ~ if (is.null(.x)) NA_real_ else .x$conf.int[1] * sqrt(2)),
cor_ars_n_ci_ub = map_dbl(ars_n_cor_test, ~ if (is.null(.x)) NA_real_ else .x$conf.int[2] * sqrt(2))
) %>%
select(-lrs_cor_test, -ars_n_cor_test) %>% # drop the list‑columns once unpacked
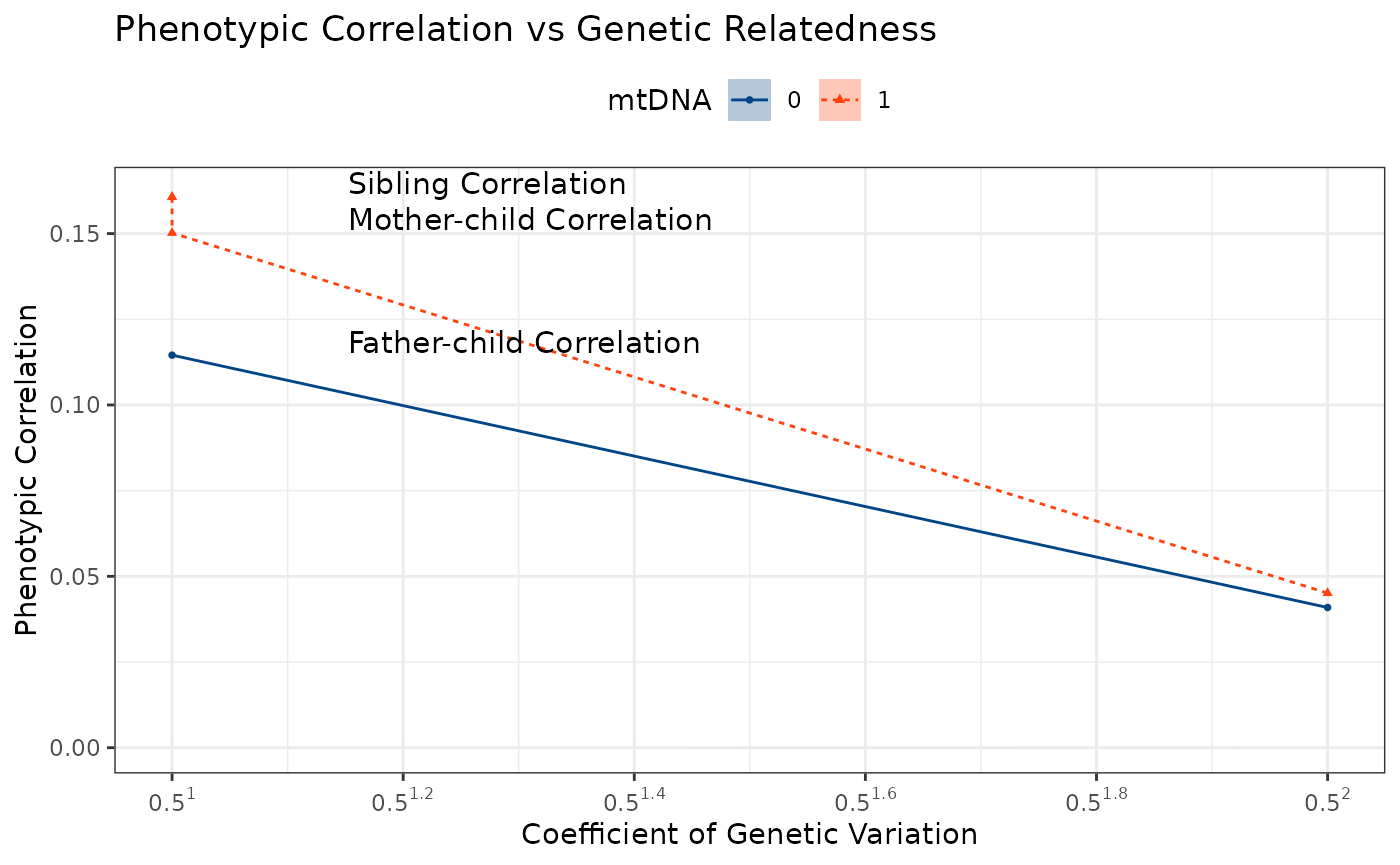
rename(cnu = cnuRel, mtdna = mitRel)The resulting data frame result contains the phenotypic
correlations for lifetime reproductive success (LRS) and annual
reproductive success (ARS-n) across different degrees of relatedness,
along with confidence intervals and p-values.
head(result)
#> # A tibble: 6 × 20
#> addRel_factor mtdna cnu n_pairs addRel_mean addRel_sd addRel_min addRel_max
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 addRel_1 1 1 103 1 1.90e-17 1 1
#> 2 addRel_0.5 0 0 1 0.5 0 0.5 0.5
#> 3 addRel_0.5 1 0 101 0.5 2.35e-17 0.5 0.5
#> 4 addRel_0.25 0 0 1 0.25 0 0.25 0.25
#> 5 addRel_0.25 1 0 544 0.25 1.43e-17 0.25 0.25
#> 6 addRel_0.125 0 0 8 0.125 0 0.125 0.125
#> # ℹ 12 more variables: cor_lrs <dbl>, cor_lrs_stat <dbl>, cor_lrs_p <dbl>,
#> # cor_lrs_df <dbl>, cor_lrs_ci_lb <dbl>, cor_lrs_ci_ub <dbl>,
#> # cor_ars_n <dbl>, cor_ars_n_stat <dbl>, cor_ars_n_p <dbl>,
#> # cor_ars_n_df <dbl>, cor_ars_n_ci_lb <dbl>, cor_ars_n_ci_ub <dbl>Phenotypic Correlation by Degree of Relatedness
ggPhenotypeByDegree(
df = result,
y_var = "cor_lrs",
y_ci_lb = "cor_lrs_ci_lb",
y_ci_ub = "cor_lrs_ci_ub",
config = list(
use_only_classic_kin = FALSE,
drop_classic_kin = FALSE,
group_by_kin = TRUE,
use_relative_degree = TRUE,
drop_non_classic_sibs = FALSE,
filter_degree_max = 12,
grouping_column = "mtdna_factor",
filter_n_pairs = 10
)
)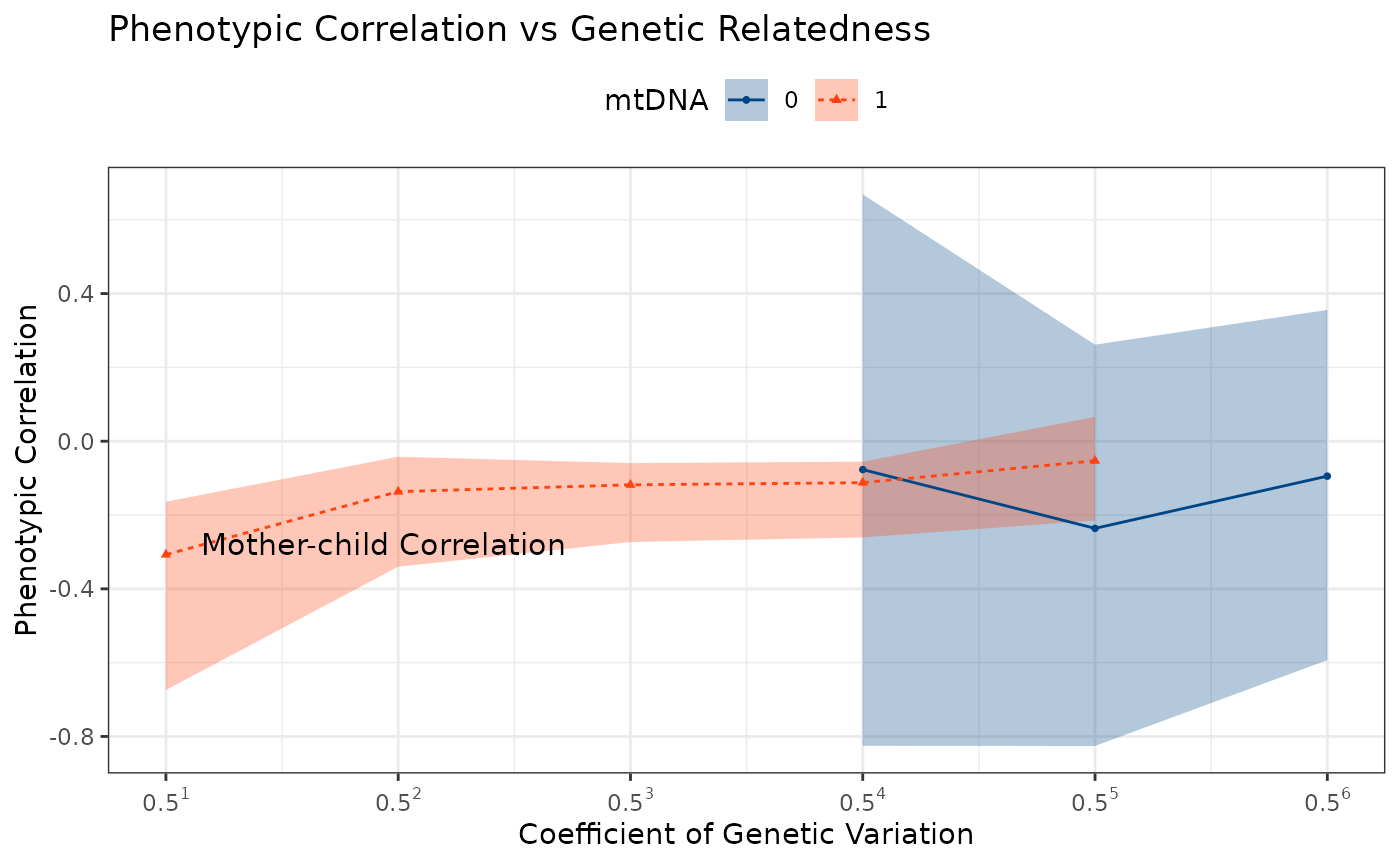
Pedigree Setup
Click to expand pedigree setup
library(tibble)
library(dplyr)
library(ggpedigree)
df <- pedigree_df <- tribble(
~n_pairs,
~addRel_min,
~addRel_max,
~addRel_emp_min,
~addRel_emp_mean,
~addRel_emp_median,
~addRel_emp_max,
~mtdna,
~cnu,
~age_k1_meanFunction,
~male_k1_meanFunction,
~same_matID_meanFunction,
~same_patID_meanFunction,
~USA_flag_10_k1_meanFunction,
~USA_flag_10_polychorFunction_rho,
~USA_flag_10_polychorFunction_se,
~USA_flag_10_polychorFunction_chisq,
~USA_flag_10_polychorFunction_df,
~USA_flag_10_ml_polychorFunction,
3617250, 0.225, 0.275, 0.2255, 0.2477, 0.25, 0.2749, 1, 0, 59.2335, 0.4806, NA, NA, 0.2497, 0.0451, 0.0006, 0.00000799, 0, 0.0451,
4983424, 0.225, 0.275, 0.2255, 0.2479, 0.25, 0.2749, 0, 0, 60.2924, 0.5468, NA, NA, 0.2470, 0.0409, 0.0005, 0.00000799, 0, 0.0409,
120024, 0.275, 0.45, 0.359375, 0.42720031928978, 0.4375, 0.4492, 1, 1, 55.3786, 0.519857671410953, NA, NA, 0.2082, 0.1692, 0.00393269696620211, 0.0000081, 0, 0.169234306738862,
137947, 0.275, 0.45, 0.2751, 0.3368, 0.34375, 0.4472, 1, 0, 58.7404, 0.4364, NA, NA, 0.230130064360623, 0.0979941906533943, 0.00359788288853913, 0.00000800052657723427, 0, 0.0979945954952227,
108989, 0.275, 0.45, 0.275390625, 0.328092281014401, 0.3125, 0.4453125, 0, 0, 60.2531852003308, 0.619355993501189, NA, NA, 0.233078768161892, 0.0774003012451702, 0.00404657995476565, 0.00000800139969214797, 0, 0.0774005589379321,
1668205, 0.45, 0.55, 0.4501953125, 0.495319721631096, 0.4990234375, 0.549560546875, 1, 1, 57.9295757847709, 0.522333747318167, NA, NA, 0.257514897453126, 0.160645710133276, 0.000975187176863543, 0.0000904696062207222, 0, 0.16065612400207,
729441, 0.45, 0.55, 0.451171875, 0.496536543871464, 0.5, 0.5498046875, 1, 0, 66.1279781018233, 0.275059425002599, NA, NA, 0.265100488202374, 0.150191012774079, 0.00152996759330951, 0.00000815372914075851, 0, 0.150193105587773,
747643, 0.45, 0.55, 0.451171875, 0.49663045074013, 0.5, 0.5478515625, 0, 0, 65.9430606759445, 0.749751203807075, NA, NA, 0.262018010446397, 0.114532502819044, 0.00154238295730097, 0.00000800378620624542, 0, 0.114533380385551,
44787, 0.55, 0.9, 0.55029296875, 0.724711065168643, 0.75, 0.8984375, 1, 1, 63.4350717031309, 0.520855662736692, NA, NA, 0.232835687466476, 0.9999, Inf, 29160.6932985728, 0, 0.952963916809192,
4639, 0.55, 0.9, 0.55029296875, 0.569994330072125, 0.5625, 0.75, 1, 0, 62.3524143288736, 0.366174409830764, NA, NA, 0.231225722917566, 0.12373864513677, 0.0194548428304584, 0.00000800392444944009, 0, 0.123740641721645,
2744, 0.55, 0.9, 0.55078125, 0.566952741528391, 0.5625, 0.75, 0, 0, 62.2241888686131, 0.758432087511395, NA, NA, 0.228467153284672, 0.123400997830296, 0.0254117516477019, 0.00000801651367510203, 0, 0.123405021390264,
1018929, 0.9, 1.1, 0.90234375, 0.994306976854356, 1, 1.09375, 1, 1, 61.9059012997516, 0.523600756854653, NA, NA, 0.254706541475618, 0.9999, Inf, 12530.2189094715, 0, 0.9999,
352, 1.1, 1.5, 1.1005859375, 1.1284, 1.125, 1.25, 1, 1, 47.7579829059829, 0.545454545454545, NA, NA, 0.153846153846154, 0.9999, Inf, 3.76616019073219, 0, 0.9999
)
ggPhenotypeByDegree(
df = df,
y_var = "USA_flag_10_polychorFunction_rho",
y_stem_se = "USA_flag_10_polychorFunction",
y_se = "USA_flag_10_polychorFunction_se"
)